Demo: Run a Full Analysis Flow
Overview
Teaching: 20 min
Exercises: 45 minQuestions
How do I follow the progress of a workflow?
What are the different steps in the example workflow?
Objectives
Mimic a full analysis flow
Understand structure of a workflow
Find the recid for a dataset and put it into the yaml file
Follow the workflow progress
You can watch the progress of the workflow either on the command line or in the Argo GUI.
On the command line, you can see the state of the workflow with
argo get @latest -n argo
and you can check the logs with
argo logs @latest -n argo
You can also get the logs from the pods (take the pod name from the listing of argo get... output) using
kubectl logs <pod-name> -n argo
You check the status of the pod
kubectl describe pod <pod-name> -n argo
When the workflow has finished, you will be able to access the output files from the http file server. You can also see the contents of the disk with
kubectl exec pv-pod -n argo -- ls /mnt/data
Expanding the Yaml File
In the previous section, you downloaded a workflow definition and submitted it. It should now be running. This workflow corresponds to the analysis example presented in this workshop.
The workflow mimicks a full analysis, first processing CMS open data samples with POET and then running an analysis script on the output files.
Open up the file argo-poet.yaml, and take a look through its contents. Below is an explanation of the major steps.
The workflow calls and runs multiple different tasks. The file is broken up into different templates. Note that each task runs in a container, and the workflow is using the same container images that we have been using in the workshop.
-
cms-od-example, the first template, is the entrypoint, and it contains the outline for the rest of the workflow. The listing under
dagdefines the inputs and outputs of each step and their dependencies. -
prepare-template gets the directories ready for other workflow steps.
-
get-metadata-template uses
cernopendata-clientto get the metadata of the dataset. -
joblist-template prepares an array that the next step would iterate over.
-
runpoet-template processes the data, and it will take the longest amount of time to complete.
-
merge-step-template combines the inputs from the jobs of the previous steps into a single file.
-
analysis-step-template creates some histograms to check that the processing went OK.
The “runpoet” step takes the array of the preceding step as input and iterates over it. It runs multiple jobs at the same time. The Argo GUI helps us visualize this process.
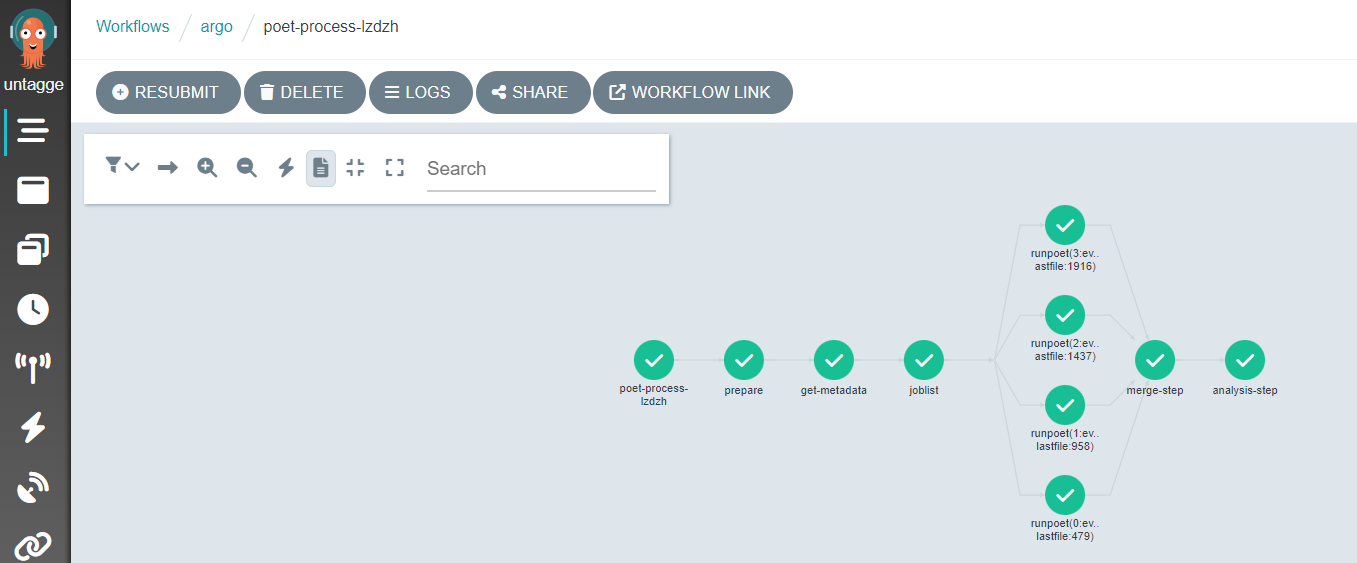
Depending on the resources you allocate to your cluster, there is a limit to the number of pods you have running at one time. If you have more pods than this number, they will wait for each other to complete.
Workflow input parameters
The workflow takes the following parameters:
arguments:
parameters:
- name: startFile
value: 1
- name: nEvents
value: 10000
- name: recid
value: 24119
- name: nJobs
value: 4
and they need to be defined as part of the first template:
templates:
- name: cms-od-example
inputs:
parameters:
- name: startFile
- name: nEvents
- name: recid
- name: nJobs
They give input to the workflow steps.
startFileis the first file in the list to be processednEventsis the number of events to be processedrecidis the dataset to be processed.nJobsis the number of separate jobs
This implementation is mainly for small-scale testing but in principle can be run with any number of events and jobs.
Getting metadata
The metadata are retrieved using the cernopendata-client container image. It is available also as a command-line tool. Task get-metadata make the following queries:
cernopendata-client get-file-locations --recid "" --protocol xrootdfor a listing of all files in a datasetcernopendata-client get-metadata --recid "" --output-value type.secondarythe type of data (Collision/Simulated)cernopendata-client get-metadata --recid "" --output-value distribution.number_filesfor number of files in the dataset.
Leaving out the --output-value option would give all metadata, which could also be inspected directly from the open data portal records by adding /export/json to the record URL.
Passing information from one task to another
The main challenge in any workflow language is the communication between the tasks. This workflow implementation illustrates some of the possibilities when using Argo as a workflow language:
- a mounted volume
/mnt/vol, available as apersistent volumeto all tasks - used for files-
the persistent volume claim is defined in the beginning with
volumes: - name: task-pv-storage persistentVolumeClaim: claimName: nfs-<NUMBER> -
it can then be used in those steps that need access to it with
volumeMounts: - name: task-pv-storage mountPath: /mnt/vol
-
- input parameters - used for configurable input parameters
-
they are defined for each step in the
dagsection, e.g.- name: get-metadata dependencies: [prepare] template: get-metadata-template arguments: parameters: - name: recid value: "{{inputs.parameters.recid}}" -
and later in the workflow in the step implementation
- name: get-metadata-template inputs: parameters: - name: recid
-
- output parameters - used to pass the output from one task to another through a defined parameter
-
they are defined in the
dagsection, e.g.- name: dataType value: "{{outputs.parameters.dataType}}" -
and in the step implementation in which they must have a default value:
outputs: parameters: - name: dataType valueFrom: default: "default" path: /tmp/type.txt
-
- output to stdout - used to pass the stdout output of one task to another.
Getting the output
Now back to the workflow that you submitted earlier. When it has completed, you will see the output in the http file-server browser window. Please note that each download costs money so do not download big files repeatedly for this workshop hands-on exercise.
Once the workflow is complete, make sure to delete it so that the cluster can scale down and does not consume resources unnecessarily:
argo delete @latest -n argo
This will not delete the output files as they were written to a persistent disk, but it removes all the pods on which tasks were running. Note that if you run the workflow again, it will overwrite the files of the previous run.
Warning!
Cloud Shell has a usage limit of 50 hours per week. In order to stay within that limit, take the habit of closing Cloud Shell as soon as you are done.
If you need more time, please install the
gcloudCLI locally, and connect to the cluster through it from your local terminal. The connect command is the same that you would use to connect in Cloud Shell.
Remarks
This is an example workflow for demonstration purposes. To keep it simple, it does not include any error handling or extensive bookkeeping. It is, however, a typical processing task that we envisage CMS open data users to be interested in.
We have implemented this example in Argo. There are many other workflow languages available, and Argo is only one of them.
Key Points
You can run an analysis on an entire dataset using multiple templates and scattering