Write the code
Last updated on 2024-07-30 | Edit this page
Estimated time: 32 minutes
Overview
Questions
- How do you write the code that matches our physics selection criteria?
- How do we keep track of everything?
- What do we write out when we process a file?
Objectives
- Learn to use
awkwardarrays to select subsets of the data - Understand how to apply the luminosity mask
- Make a first-order plot of some of our variables of interest
- Process at least one file of both simulation and collision data
Introduction
There is a significant amount of code to run here and so we have written the majority of it in a Jupyter notebook. You can run most of the code on its own, but you should take the time to read and understand what is happening. In some places, you need to modify the code to get it to run.
First, start your python docker container, following the lessons from
the
pre-exercises. I am on a Linux machine, and I have already created
the cms_open_data_python directory. So I will do the
following
Start the container with
BASH
docker run -it --name my_python -P -p 8888:8888 -v ${workpath}/cms_open_data_python:/code gitlab-registry.cern.ch/cms-cloud/python-vnc:python3.10.5You will get a container prompt similar this:
OUTPUT
cmsusr@4fa5ac484d6f:/code$Before we start our Jupyter environment, let’s download the notebook we’ll be using with the following command.
BASH
wget https://raw.githubusercontent.com/cms-opendata-workshop/workshop2024-lesson-event-selection/main/instructors/data_selection_lesson.ipynbNow I will start Jupyter lab as follows.
and open the link that is printed out in the message.
How to follow this lesson
While some of the code will be explained on this web page, the majority of the code and explanations of the code are written out in the Jupyter notebook. Therefore, you should primarily following along there.
I will use this webpage for the lesson to provide guideposts and checkpoints that we can refer to as we work through the lesson.
Running through the selection steps (in the Jupyter notebook)
Preparing the environment
We will be making extensive use of the uproot and
awkward python libraries, the pandas data
library, and a few other standard python libraries. The first part of
the notebook Install and upgrade libraries asks you to
do just that, in order to ensure a consistent environment.
Depending on your connection, it should take 1-2 minutes to upload and import the libraries.
We’ve also prepared some helper code that makes it easier to work with the data in this lesson. You can see the code here but we will explain the functions and data objects in this notebook.
Read in some files
Run through the notebook for the Download essential files section.
How many input NanoAOD files will we process?
How many collision files are there? How many signal files are there? How many files are there combined in the background sample files?
You will run
And get
OUTPUT
68 FILE_LIST_Wjets.txt
152 FILE_LIST_collision.txt
4 FILE_LIST_signal_M2000.txt
146 FILE_LIST_tthadronic.txt
49 FILE_LIST_ttleptonic.txt
138 FILE_LIST_ttsemilep.txt
557 totalSo there are 152 files in the collision dataset.
4 files in our signal_M2000 dataset.
If we add up the background samples of ttXXX and
Wjets we find there are 401 total files.
Apply the cuts
Run through the next sections in the notebook to set up the cuts.
Check your understanding as you go.
Reconstruct the resonance candidate mass
To reconstruct the \(z'\) candidate mass in a computationally-efficient way, we are going to make use of the Vector library, which works very well with awkward, as you can see in these examples.
It allows for very simple code that will automatically calculate all combinations of particles when reconstructing some parent candidate.
Plot
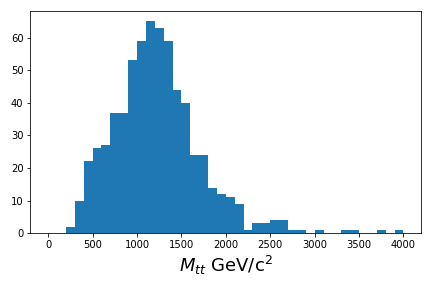
If you are able to run though Use the cuts and calculate some values, you should have been able to produce a basic plot.
Plot the ttbar mass
Key Points
- Awkward arrays allow for a simplified syntax when making cuts to select the data
- You need to be careful to distinguish between cuts on events and cuts on particles