Introduction
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How can we set up an environment to run over Run I simplified files.
Objectives
Remind ourselves of how to set up Docker to use ROOT.
Setting up your environment
Using a local environment
If you already have an environment set up locally on your computer that has ROOT version 6 or higher, you should be able to run these exercises without any additional changes.
Using Docker
If you do not have a completely local installation of ROOT, you are recommended to use Docker, as per the pre-exercises and previous lessons.
Ideally, you have already downloaded Docker and tested it out when you completed the Docker pre-exercise for this workshop.
The specific episode is Using Docker with CMS Open Data. You will want to scroll down to Download the docker images for ROOT and python tools and start container and then just below that, ROOT container. Please make sure you have gone through that lesson, as we will be following most of the instructions about how to launch the container, with some minimal modifications.
Create a local directory to store your work
Before you start up the Docker container, create a local directory where we will store
files. Let’s call it cms_open_data_run1. If you were on Linux or in a Mac terminal,
you would type something like
mkdir cms_open_data_run1
Launch Docker
We’ll follow the instructions from the previous lesson on Docker except that
- We will call this new instance of the container
my_run1 - We will mount a new
volumewhich is ourcms_open_data_run1directory
If you are on native Linux and want to use X11-forwarding, use
docker run -it --name my_run1 --net=host --env="DISPLAY" -v $HOME/.Xauthority:/home/cmsusr/.Xauthority:rw -v ${HOME}/cms_open_data_run1:/code gitlab-registry.cern.ch/cms-cloud/root-vnc:latest
On MacOS and Windows WSL2 (and on native Linux if you do not want to use X11-forwarding), use
docker run -it --name my_run1 -P -p 5901:5901 -p 6080:6080 -v ${HOME}/cms_open_data_run1:/code gitlab-registry.cern.ch/cms-cloud/root-vnc:latest
This opens a bash shell where you can type your commands. Edit files in the cms_open_data_run1 directory on your local computer, but run the commands in the container.
For graphics, on native Linux, use X11-forwarding. On other systems, use VNC that is installed in the container and start the graphics windows with vnc_start. Open the browser window in the address given at the start message (http://127.0.0.1:6080/vnc.html) with the default VNC password is cms.cern. It shows an empty screen to start with and all graphics will pop up there.
Type exit to leave the container, and if you have started VNC, stop it first:
stop_vnc
exit
Key Points
Docker can reduce the overhead in setting up a ROOT environment.
Analyzing Run 1 data
Overview
Teaching: 10 min
Exercises: 50 minQuestions
How can I analyze larger ROOT files?
Objectives
Demonstrate examples of accessing ROOT files remotely
To compare and contrast using standard ROOT approaches and newer ROOT objects like RDataFrame
Analyzying the dimuon samples
This lesson will be primarily following the material found here about using the NanoAOD for Run 1 format in an analysis of the dimuon samples.
Potential pitfalls!
We’ll be running over some larger ROOT files in this lesson and for some of you, memory issues may cause some errors or crashes of the code. If that happens, it is primarily restricted to this exercise and you should feel free to simply follow along with the instructor.
Download the code and scripts
Launch your Docker container, as per the previous episode.
From inside your Docker container, we’re going to execute a series of curl commands.
curl is a widely used utility to download files from remote locations.
Simply highlight the commands below and cut-and-paste them into your Docker terminal, or your
local terminal if you are working without Docker.
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/Dimuon2011_eospublic.C --output Dimuon2011_eospublic.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/Dimuon2011_eospublic_RDF.C --output Dimuon2011_eospublic_RDF.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/Dimuon2011_local.C --output Dimuon2011_local.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/Dimuon2011_local_RDF.C --output Dimuon2011_local_RDF.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/MuHistos_eospublic.cxx --output MuHistos_eospublic.cxx
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/MuHistos_local.cxx --output MuHistos_local.cxx
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/dimuonSpectrum2012_eospublic.C --output dimuonSpectrum2012_eospublic.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/dimuonSpectrum2012_eospublic_test.C --output dimuonSpectrum2012_eospublic_test.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/dimuonSpectrum2012_eospublic.py.txt --output dimuonSpectrum2012_eospublic.py
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/dimuonSpectrum2012_local.C --output dimuonSpectrum2012_local.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/dimuonSpectrum2012_local.py.txt --output dimuonSpectrum2012_local.py
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/dimuonSpectrum2012_outreach.C --output dimuonSpectrum2012_outreach.C
curl https://twiki.cern.ch/twiki/pub/CMSPublic/NanoAODRun1Examples/dimuonSpectrum2012_outreach.py.txt --output dimuonSpectrum2012_outreach.py
You can look to see that the files were downloaded properly by typing
ls -ltr
Run some of the commands
Your instructor will be executing most of these commands, some of which might take a few minutes to run.
In most cases, these scripts produce an output .pdf file with a name similar to that of the script itself.
If you have any issues with ROOT displaying the plots as they are made, you can view the .pdf files
on your local system by looking in the cms_open_data_run1 directory that you created.
Zeroth example
The first thing we will do is run a very simple ROOT script that loads in a single, relatively small file from your laptop and produces a plot of the dimuon spectrum. Because it is a single file, we will not have the same amount of data in this plot as later examples, but you can use this to check your connection and X windows forwarding (e.g. does the plot pop up?).
First, let’s download the file, using one of the XRootD utilities,
xrdcp that allows us to download files.
xrdcp root://eospublic.cern.ch//eos/opendata/cms/upload/NanoAODRun1/01-Jul-22/Run2012B_DoubleMuParked/01-Jul-22Run2012B_DoubleMuParked/03C5684F-8BAF-4312-8235-2B0039F2FB93.root .
The file is about 1.4 Gb, but should take less than a minute to download.
xrdcpalternativeIf
xrdcpis not running/working, you can also download this file with this commandcurl https://opendata.cern.ch/record/6004/files/assets/cms/upload/NanoAODRun1/01-Jul-22/Run2012B_DoubleMuParked/01-Jul-22Run2012B_DoubleMuParked/03C5684F-8BAF-4312-8235-2B0039F2FB93.root --output 03C5684F-8BAF-4312-8235-2B0039F2FB93.root
Once it it is downloaded, you can process this one file by running dimuonSpectrum2012_eospublic_test.C in ROOT.
To do so, you will launch ROOT with the name of the script as an argument.
root -l dimuonSpectrum2012_eospublic_test.C
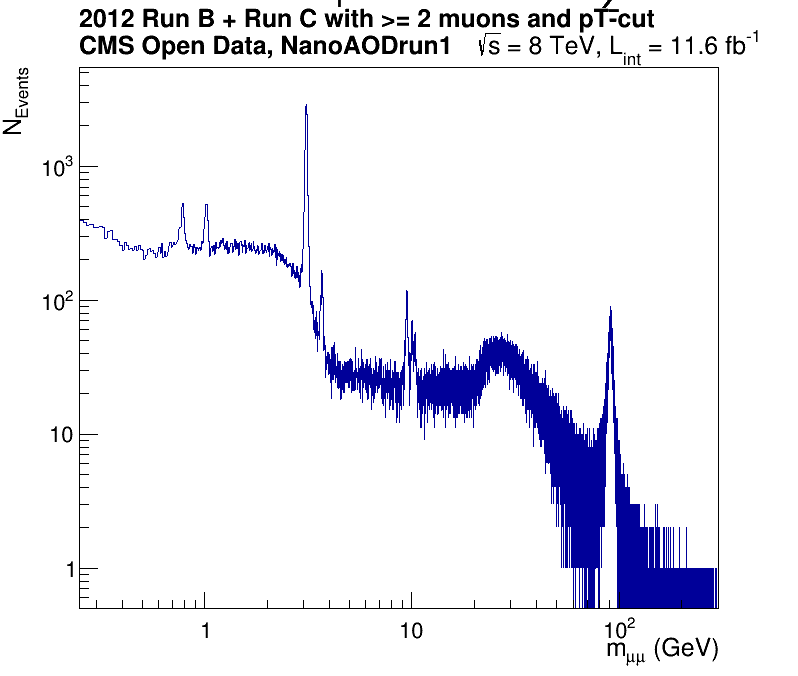
It should take less than one minute to run and if it does, you will see a window pop up that looks like the following image.
CMS dimuon spectrum - 2012 data sample
Invariant mass of a select sample of oppositely charged dimuon pairs. Derived from a smaller subset of 2012 data.
If you are having issues with X11 forwarding, the script should still create a file
dimuonSpectrum2012_C_eospublic.pdf in the cms_open_data_run1 directory you made,
and you can view it there.
In the following sections, the scripts are written so as to run over larger files that you access remotely. Depending on your connection, it may take longer than the time alotted for this activity during the workshop, in which case you are encouraged to follow along with the instructor and run these on your own time, if you so choose.
First example
Let’s run the first command making use of ROOT’s ability to compile and execute a file in one step. This might take 5 minutes for local participants at CERN, but longer for remote participants.
root -l MuHistos_eospublic.cxx++
root [0]
Processing MuHistos_eospublic.cxx++...
Info in <TUnixSystem::ACLiC>: creating shared library /code/./MuHistos_eospublic_cxx.so
reading root://eospublic.cern.ch//eos/opendata/cms/upload/NanoAODRun1/01-Jul-22/Run2010B_Mu_merged.root
writing to MuHistos_Mu_eospublic.root
entries = 26718043
event nr 0
event nr 1000000
event nr 2000000
event nr 3000000
event nr 4000000
event nr 5000000
event nr 6000000
event nr 7000000
event nr 8000000
event nr 9000000
event nr 10000000
event nr 11000000
event nr 12000000
event nr 13000000
event nr 14000000
event nr 15000000
event nr 16000000
event nr 17000000
event nr 18000000
event nr 19000000
event nr 20000000
event nr 21000000
event nr 22000000
event nr 23000000
event nr 24000000
event nr 25000000
event nr 26000000
After the above output, the program will finish and will return the command-line prompt.
The program should have produced an output file called MuHistos_Mu_eospublic.root. You can check this by typing
ls -l MuHistos_Mu_eospublic.root
-rw-r--r-- 1 cmsusr cmsusr 31625 Jul 31 22:57 MuHistos_Mu_eospublic.root
You can open this file in ROOT and inspect it with a TBrowser. First type
root -l MuHistos_Mu_eospublic.root
This will put you into the ROOT environment, from which you can then launch the TBrowser
from the prompt (you needn’t type the root [0]).
root [0] TBrowser b;
You can then click on the file name in the window and then click on the various histograms to view them.
Example 2 (RDataFrame)
A different example runs over a smaller (2 Gb) file, primarily used for
outreach, and makes use of ROOT’s relatively
newer RDataFrame object. You can run this example by launching ROOT from the commandline.
root -l dimuonSpectrum2012_outreach.C
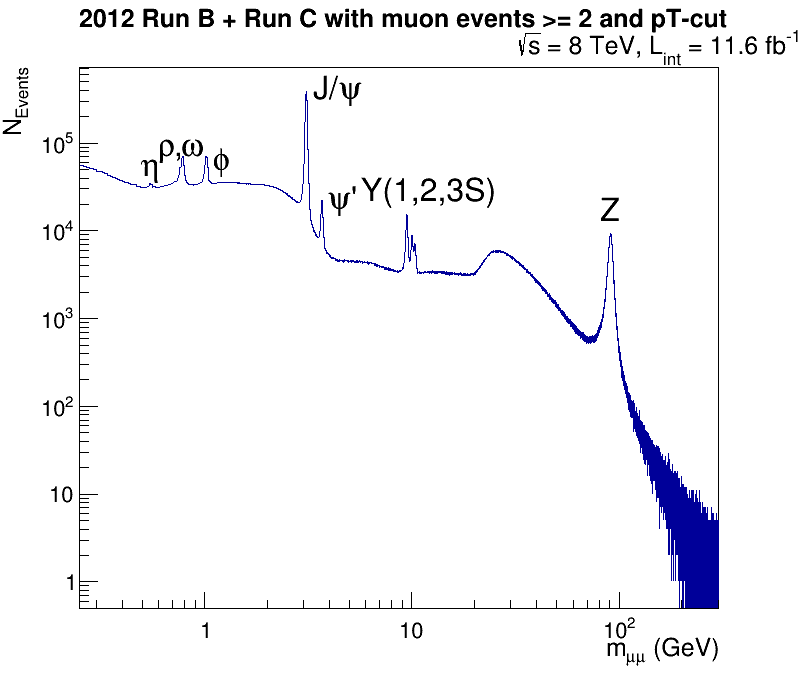
It should only take a few minutes to run and if X-forwarding is working for you, you should see a ROOT window pop up that looks like this.
CMS dimuon spectrum
Invariant mass of a select sample of oppositely charged dimuon pairs.
Example 3 (2011 data)
This example runs over 2011 data that was used for more “real” analysis. Takes about 15 minutes at CERN.
root -l Dimuon2011_eospublic_RDF.C
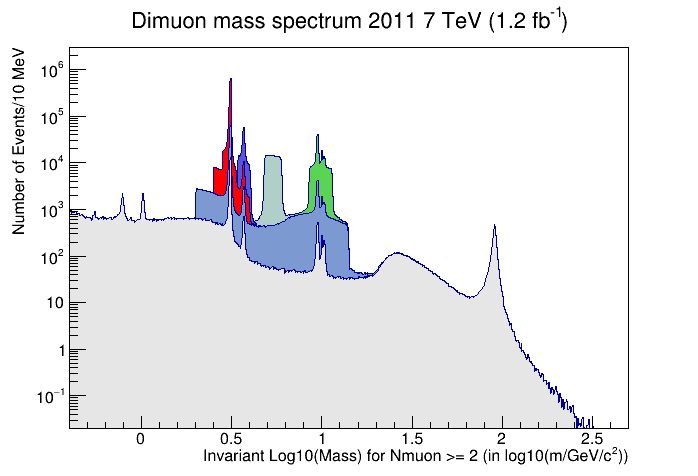
When it finishes, it should pop up a window with the following plot.
CMS dimuon spectrum - 2011 data sample
Invariant mass of a select sample of oppositely charged dimuon pairs. Derived from 2011 data.
Key Points
Making use of RDataFrame can speed up your analysis
You have different options to call ROOT
You can access files remotely or download them for local access