Introduction
Overview
Teaching: 5 min
Exercises: 10 minQuestions
How do I make plots of the skimmed datasets?
How do I setup my ROOT environment?
Objectives
Set up a ROOT environment that works for you
Launch ROOT to test the setup
Quickly browse the contents of a ROOT file with TBrowser
So you’ve skimmed some data and you want to start looking at it. Great! Since these are most likely ROOT files, it’s worth starting with ROOT to inspect them and make some plots. There are other standalone tools like uproot that allow you to interact with these files, but we’ll stick with ROOT for this exercise.
Setting up a ROOT environment
Ideally, prior to starting this exercise, you will have gone through the ROOT pre-exercise. If you haven’t gone through the full tutorial, that’s OK, but you should definitely have gone through the module that shows you how to setup a working ROOT environment.
Workflow for this lesson
We have found that there are sometimes issues with X11 forwarding and getting windows and GUIs to pop up when the applications are launched from within docker or a VM. That’s OK!
In this lesson, you can choose to view the plots one of two different ways:
- If X11 forwarding works, you will see some of the plots pop up.
- Otherwise, the scripts we’ll be using will produce
.pngimages which you can copy to your local machine for viewing.It’s up to you which approach you want to use.
You will need to edit the files though, so whichever workflow you use, make sure you have an editor you are comfortable with.
Docker-specific commands for X11 forwarding and other miscellany
If you want to use same ROOT-installed docker container as the pre-exercises and want to have X11 forwarding and a named container so you can come back to it, you can run the following command:
docker run --name root6 --net=host --env="DISPLAY" --volume="$HOME/.Xauthority:/home/cmsusr/.Xauthority:rw" -it rootproject/root:6.22.02-conda /bin/bash
If you’re running the above for the first time, it may take ~10 minutes to download the Docker files.
Afterwards, you can alway restart the container with
docker start -i root6
Note that when the container starts up, you are in the / directory. You will want to
get to the home directory
/root by typing
cd
You should make sure there is an editor you feel comfortable with. You can install
new packages with apt. For example, to install vim or nano you would run one of
the following from inside the docker container.
apt install vim
apt install nano
VM-specific commands go here
Looking forward to some feedback from the participants! :)
Download the files and code we’ll use
We are going to be running through parts of the open data analysis, Analysis of Higgs boson decays to two tau leptons using data and simulation of events at the CMS detector from 2012, which some of you may have done as part of the pre-exercises.
Analysis description
This analysis uses data and simulation of events at the CMS experiment from 2012 with the goal to study decays of a Higgs boson into two tau leptons in the final state of a muon lepton and a hadronically decayed tau lepton. The analysis follows loosely the setup of the official CMS analysis published in 2014.
We’ll be looking at the skimmed ROOT files, but those files can take considerable time to generate, depending on where you are and your internet connection. To save time, we’ve run Step 1 of that exercise already and generated the skimmed ROOT files. We’ve also modified some of the scripts slightly to make it easier to copy the files out of the VM/container and included some new, simpler files that allow you to inspect these ROOT files and make some first-order plots.
If you haven’t yet, you may want to check out the analysis to get a feel for the physics.
You can download the files and code by checking out the master branch of this lesson.
cd
git clone -b master git://github.com/cms-opendata-workshop/workshop-lesson-plotting-and-interpretation
This can take a few minutes as it’s about 60 MB of files.
Once it’s downloaded, you can descend into the appropriate directory.
We’ll also want to make a plots directory here. The set of commands
is given below.
cd workshop-lesson-plotting-and-interpretation/scripts_and_data
mkdir plots
ls -l
-rw-r--r-- 1 root root 7524470 Oct 1 05:20 DYJetsToLLSkim.root
-rw-r--r-- 1 root root 786366 Oct 1 05:20 GluGluToHToTauTauSkim.root
-rw-r--r-- 1 root root 13548245 Oct 1 05:20 Run2012B_TauPlusXSkim.root
-rw-r--r-- 1 root root 20688325 Oct 1 05:20 Run2012C_TauPlusXSkim.root
-rw-r--r-- 1 root root 4769020 Oct 1 05:20 TTbarSkim.root
-rw-r--r-- 1 root root 1156105 Oct 1 05:20 VBF_HToTauTauSkim.root
-rw-r--r-- 1 root root 3591969 Oct 1 05:20 W1JetsToLNuSkim.root
-rw-r--r-- 1 root root 5965972 Oct 1 05:20 W2JetsToLNuSkim.root
-rw-r--r-- 1 root root 3520631 Oct 1 05:20 W3JetsToLNuSkim.root
-rw-r--r-- 1 root root 3048 Oct 1 05:20 compare_two_files.py
-rw-r--r-- 1 root root 3264 Oct 1 05:20 dump_and_plot.py
-rw-r--r-- 1 root root 5864 Oct 1 05:20 histograms.py
-rw-r--r-- 1 root root 7655 Oct 1 05:20 plot.py
drwxr-xr-x 2 root root 4096 Oct 1 05:24 plots
Testing ROOT
From this directory, lets try opening one of these files, the signal Monte Carlo,
GluGluToHToTauTauSkim.root.
We can do this from the ROOT C-interpreter.
root GluGluToHToTauTauSkim.root
------------------------------------------------------------------
| Welcome to ROOT 6.22/02 https://root.cern |
| (c) 1995-2020, The ROOT Team; conception: R. Brun, F. Rademakers |
| Built for linuxx8664gcc on Aug 18 2020, 07:08:00 |
| From tag , 17 August 2020 |
| Try '.help', '.demo', '.license', '.credits', '.quit'/'.q' |
------------------------------------------------------------------
root [0]
Attaching file GluGluToHToTauTauSkim.root as _file0...
(TFile *) 0x56356e5cf260
Testing X11 forward (for some users)
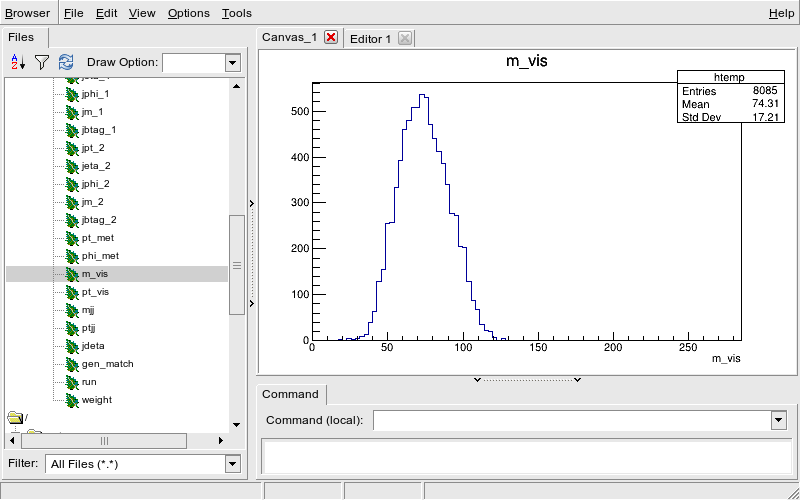
If you have been able to enable X11 forwarding, you can launch the ROOT TBrowser to inspect the ROOT file and see what is in there.
Assuming you have run the previous ROOT command and are in the CINT, you can now
root [1] TBrowser b;And you should see the TBrowser pop up.
Double-click on the
GluGluToHToTauTauSkim.rootand thenEventsand you can browse the entries and make some histograms.
Whether or not you have gotten X11 working, you can exit ROOT by typing .q.
root [2] .q
Key Points
The most straightforward way to open your skimmed files is with ROOT
There are different plotting libraries out there but you can get started pretty easily with ROOT