Introduction
Overview
Teaching: 30 min
Exercises: 0 minQuestions
What is Kubernetes?
Why would I use Kubernetes?
Objectives
Get a rough understanding of Kubernetes and some of its components
The introduction will be performed using slides available on the Workshop Indico page.
Once you have successfully completed the steps there, please continue to the next episode.
Key Points
Kubernetes is a powerful tool to schedule containers.
Getting started with Google Kubernetes Engine
Overview
Teaching: 15 min
Exercises: 15 minQuestions
How to create a cluster on Google Cloud Platform?
How to setup Google Kubernetes Engine?
How to setup a workflow engine to submit jobs?
How to run a simple job?
Objectives
Understand how to run a simple workflows in a commercial cloud environment
Get access to Google Cloud Platform
Usually, you would need to create your own account (and add a credit card) in order to be able to access the public cloud. For this workshop, however, you will have access to Google Cloud via the ARCHIVER project. Alternatively, you can create a Google Cloud account and you will get free credits worth $300, valid for 90 days (requires a credit card).
The following steps will be performed using the Google Cloud Platform console.
Use an incognito/private browser window
We strongly recommend you use an incognito/private browser window to avoid possible confusion and mixups with personal Google accounts you might have.
Find your cluster
By now, you should already have created your first Kubernetes cluster. In case you do not find it, use the “Search products and resources” field to search and select “Kubernetes Engine”. It should show up there.
Open the working environment
You can work from the cloud shell, which opens from an icon in the tool bar, indicated by the red arrow in the figure below:

In the following, all the commands are typed in that shell.
We will make use of many kubectl commands. If interested, you can read
an overview
of this command line tool for Kubernetes.
Install argo as a workflow engine
While jobs can also be run manually, a workflow engine makes defining and submitting jobs easier. In this tutorial, we use argo. Install it into your working environment with the following commands (all commands to be entered into the cloud shell):
kubectl create ns argo
kubectl apply -n argo -f https://raw.githubusercontent.com/argoproj/argo/stable/manifests/quick-start-postgres.yaml
curl -sLO https://github.com/argoproj/argo/releases/download/v2.11.1/argo-linux-amd64.gz
gunzip argo-linux-amd64.gz
chmod +x argo-linux-amd64
sudo mv ./argo-linux-amd64 /usr/local/bin/argo
This will also install the argo binary, which makes managing the workflows easier.
Reconnecting after longer time away
In case you leave your computer, you might have to reconnect to the CloudShell again, and also on a different computer. If the
argocommand is not found, run the command above again starting from thecurlcommand.
You need to execute the following command so that the argo workflow controller
has sufficient rights to manage the workflow pods.
Replace XXX with the number for the login credentials you received.
kubectl create clusterrolebinding cern-cms-cluster-admin-binding --clusterrole=cluster-admin --user=cms-gXXX@arkivum.com
You can now check that argo is available with
argo version
We need to apply a small patch to the default argo config. Create a file called
patch-workflow-controller-configmap.yaml:
data:
artifactRepository: |
archiveLogs: false
Apply:
kubectl patch configmap workflow-controller-configmap -n argo --patch "$(cat patch-workflow-controller-configmap.yaml)"
Run a simple test workflow
To test the setup, run a simple test workflow with
argo submit -n argo --watch https://raw.githubusercontent.com/argoproj/argo/master/examples/hello-world.yaml
argo list -n argo
argo get -n argo @latest
argo logs -n argo @latest
argo delete -n argo @latest
Please mind that it is important to delete your workflows once they have completed. If you do not do this, the pods associated with the workflow will remain scheduled in the cluster, which might lead to additional charges. You will learn how to automatically remove them later.
Kubernetes namespaces
The above commands as well as most of the following use a flag
-n argo, which defines the namespace in which the resources are queried or created. Namespaces separate resources in the cluster, effectively giving you multiple virtual clusters within a cluster.You can change the default namespace to
argoas follows:kubectl config set-context --current --namespace=argo
Key Points
With Kubernetes one can run workflows similar to a batch system
Storing workflow output on Google Kubernetes Engine
Overview
Teaching: 15 min
Exercises: 15 minQuestions
How can I set up shared storage for my workflows?
How to run a simple job and get the the ouput?
How to run a basic CMS Open Data workflow and get the output files?
Objectives
Understand how to set up shared storage and use it in a workflow
One concept that is particularly different with respect to running on a local computer is storage. Similarly as when mounting directories or volumes in Docker, one needs to mount available storage into a Pod for it to be able to access it. Kubernetes provides a large amount of storage types.
Defining a storage volume
The test job above did not produce any output data files, just text logs. The data analysis jobs will produce output files and, in the following, we will go through a few steps to setup a volume where the output files will be written and from where they can be fetched. All definitions are passed as “yaml” files, which you’ve already used in the steps above. Due to some restrictions of the Google Kubernetes Engine, we need to use an NFS persistent volumes to allow parallel access (see also this post).
The following commands will take care of this. You can look at the content of the files that are directly applied from GitHub at the workshop-paylod-kubernetes repository.
As a first step, a volume needs to be created:
gcloud compute disks create --size=100GB --zone=us-central1-c gce-nfs-disk-<NUMBER>
Replace <NUMBER> by your account number, e.g. 023. The zone should be the
same as the one of the cluster created (no need to change here).
The output will look like this (don’t worry about the warnings):
WARNING: You have selected a disk size of under [200GB]. This may result in poor I/O performance. For more information, see: https://developers.google.com/compute/docs/disks#per
formance.
Created [https://www.googleapis.com/compute/v1/projects/cern-cms/zones/us-central1-c/disks/gce-nfs-disk-001].
NAME ZONE SIZE_GB TYPE STATUS
gce-nfs-disk-001 us-central1-c 100 pd-standard READY
New disks are unformatted. You must format and mount a disk before it
can be used. You can find instructions on how to do this at:
https://cloud.google.com/compute/docs/disks/add-persistent-disk#formatting
Now, let’s get to using this disk:
curl -LO https://raw.githubusercontent.com/cms-opendata-workshop/workshop-payload-kubernetes/master/001-nfs-server.yaml
The file looks like this:
apiVersion: apps/v1
kind: Deployment
metadata:
name: nfs-server-<NUMBER>
spec:
replicas: 1
selector:
matchLabels:
role: nfs-server-<NUMBER>
template:
metadata:
labels:
role: nfs-server-<NUMBER>
spec:
containers:
- name: nfs-server-<NUMBER>
image: gcr.io/google_containers/volume-nfs:0.8
ports:
- name: nfs
containerPort: 2049
- name: mountd
containerPort: 20048
- name: rpcbind
containerPort: 111
securityContext:
privileged: true
volumeMounts:
- mountPath: /exports
name: mypvc
volumes:
- name: mypvc
gcePersistentDisk:
pdName: gce-nfs-disk-<NUMBER>
fsType: ext4
Replace all occurences of <NUMBER> by your account number, e.g. 023.
You can edit files directly in the console or by opening the built-in
graphical editor.
Then apply the manifest:
kubectl apply -n argo -f 001-nfs-server.yaml
Then on to the server service:
curl -OL https://raw.githubusercontent.com/cms-opendata-workshop/workshop-payload-kubernetes/master/002-nfs-server-service.yaml
This looks like this:
apiVersion: v1
kind: Service
metadata:
name: nfs-server-<NUMBER>
spec:
ports:
- name: nfs
port: 2049
- name: mountd
port: 20048
- name: rpcbind
port: 111
selector:
role: nfs-server-<NUMBER>
As above, replace all occurences of <NUMBER> by your account number, e.g. 023
and apply the manifest:
kubectl apply -n argo -f 002-nfs-server-service.yaml
Download the manifest needed to create the PersistentVolume and the PersistentVolumeClaim:
curl -OL https://raw.githubusercontent.com/cms-opendata-workshop/workshop-payload-kubernetes/master/003-pv-pvc.yaml
The file looks as follows:
apiVersion: v1
kind: PersistentVolume
metadata:
name: nfs-<NUMBER>
spec:
capacity:
storage: 100Gi
accessModes:
- ReadWriteMany
nfs:
server: <Add IP here>
path: "/"
---
kind: PersistentVolumeClaim
apiVersion: v1
metadata:
name: nfs-<NUMBER>
namespace: argo
spec:
accessModes:
- ReadWriteMany
storageClassName: ""
resources:
requests:
storage: 100Gi
In the line containing server: replace <Add IP here> by the output
of the following command:
kubectl get -n argo svc nfs-server-<NUMBER> |grep ClusterIP | awk '{ print $3; }'
This command queries the nfs-server service that we created above
and then filters out the ClusterIP that we need to connect to the
NFS server. Replace <NUMBER> as before.
Apply this manifest:
kubectl apply -n argo -f 003-pv-pvc.yaml
Let’s confirm that this worked:
kubectl get pvc nfs-<NUMBER> -n argo
You will see output similar to this
NAME STATUS VOLUME CAPACITY ACCESS MODES STORAGECLASS AGE
nfs Bound nfs 100Gi RWX 5h2m
Note that it may take some time before the STATUS gets to the state “Bound”.
Now we can use this volume in the workflow definition. Create a workflow definition file argo-wf-volume.yaml with the following contents:
# argo-wf-volume.ysml
apiVersion: argoproj.io/v1alpha1
kind: Workflow
metadata:
generateName: test-hostpath-
spec:
entrypoint: test-hostpath
volumes:
- name: task-pv-storage
persistentVolumeClaim:
claimName: nfs-<NUMBER>
templates:
- name: test-hostpath
script:
image: alpine:latest
command: [sh]
source: |
echo "This is the ouput" > /mnt/vol/test.txt
echo ls -l /mnt/vol: `ls -l /mnt/vol`
volumeMounts:
- name: task-pv-storage
mountPath: /mnt/vol
Submit and check this workflow with
argo submit -n argo argo-wf-volume.yaml
argo list -n argo
Take the name of the workflow from the output (replace XXXXX in the following command) and check the logs:
kubectl logs pod/test-hostpath-XXXXX -n argo main
Once the job is done, you will see something like:
ls -l /mnt/vol: total 20 drwx------ 2 root root 16384 Sep 22 08:36 lost+found -rw-r--r-- 1 root root 18 Sep 22 08:36 test.txt
Get the output file
The example job above produced a text file as an output. It resides in the persistent volume that the workflow job has created. To copy the file from that volume to the cloud shell, we will define a container, a “storage pod” and mount the volume there so that we can get access to it.
Create a file pv-pod.yaml with the following contents:
# pv-pod.yaml
apiVersion: v1
kind: Pod
metadata:
name: pv-pod
spec:
volumes:
- name: task-pv-storage
persistentVolumeClaim:
claimName: nfs-<NUMBER>
containers:
- name: pv-container
image: busybox
command: ["tail", "-f", "/dev/null"]
volumeMounts:
- mountPath: /mnt/data
name: task-pv-storage
Create the storage pod and copy the files from there
kubectl apply -f pv-pod.yaml -n argo
kubectl cp pv-pod:/mnt/data /tmp/poddata -n argo
and you will get the file created by the job in /tmp/poddata/test.txt.
Run a CMS open data workflow
If the steps above are successful, we are now ready to run a workflow to process CMS open data.
Create a workflow file argo-wf-cms.yaml with the following content:
# argo-wf-cms.yaml
apiVersion: argoproj.io/v1alpha1
kind: Workflow
metadata:
generateName: nanoaod-argo-
spec:
entrypoint: nanoaod-argo
volumes:
- name: task-pv-storage
persistentVolumeClaim:
claimName: nfs-<NUMBER>
templates:
- name: nanoaod-argo
script:
image: cmsopendata/cmssw_5_3_32
command: [sh]
source: |
source /opt/cms/entrypoint.sh
sudo chown $USER /mnt/vol
mkdir workspace
cd workspace
git clone git://github.com/cms-opendata-analyses/AOD2NanoAODOutreachTool AOD2NanoAOD
cd AOD2NanoAOD
scram b -j8
nevents=100
eventline=$(grep maxEvents configs/data_cfg.py)
sed -i "s/$eventline/process.maxEvents = cms.untracked.PSet( input = cms.untracked.int32($nevents) )/g" configs/data_cfg.py
cmsRun configs/data_cfg.py
cp output.root /mnt/vol/
echo ls -l /mnt/vol
ls -l /mnt/vol
volumeMounts:
- name: task-pv-storage
mountPath: /mnt/vol
Submit the job with
argo submit -n argo argo-wf-cms.yaml --watch
The option --watch gives a continuous follow-up of the progress. To get the logs of the job, use the process name (nanoaod-argo-XXXXX) which you can also find with
argo get -n argo @latest
and follow the container logs with
kubectl logs pod/nanoaod-argo-XXXXX -n argo main
Get the output file output.root from the storage pod in a similar manner as it was done above.
Accessing files via http
With the storage pod, you can copy files between the storage element and the CloudConsole. However, a practical use case would be to run the “big data” workloads in the cloud, and then download the output to your local desktop or laptop for further processing. An easy way of making your files available to the outside world is to deploy a webserver that mounts the storage volume.
We first patch the config of the webserver to be created as follows:
mkdir conf.d
cd conf.d
curl -sLO https://raw.githubusercontent.com/cms-opendata-workshop/workshop-payload-kubernetes/master/conf.d/nginx-basic.conf
cd ..
kubectl create configmap basic-config --from-file=conf.d -n argo
Then prepare to deploy the fileserver by downloading the manifest:
curl -sLO https://github.com/cms-opendata-workshop/workshop-payload-kubernetes/raw/master/deployment-http-fileserver.yaml
Open this file and again adjust the <NUMBER>:
# deployment-http-fileserver.yaml
apiVersion: extensions/v1beta1
kind: Deployment
metadata:
labels:
service: http-fileserver
name: http-fileserver
spec:
replicas: 1
strategy: {}
selector:
matchLabels:
service: http-fileserver
template:
metadata:
labels:
service: http-fileserver
spec:
volumes:
- name: volume-output
persistentVolumeClaim:
claimName: nfs-<NUMBER>
- name: basic-config
configMap:
name: basic-config
containers:
- name: file-storage-container
image: nginx
ports:
- containerPort: 80
volumeMounts:
- mountPath: "/usr/share/nginx/html"
name: volume-output
- name: basic-config
mountPath: /etc/nginx/conf.d
Apply and expose the port as a LoadBalancer:
kubectl create -n argo -f deployment-http-fileserver.yaml
kubectl expose deployment http-fileserver -n argo --type LoadBalancer --port 80 --target-port 80
Exposing the deployment will take a few minutes. Run the following command to follow its status:
kubectl get svc -n argo
You will initially see a line like this:
NAME TYPE CLUSTER-IP EXTERNAL-IP PORT(S) AGE
http-fileserver LoadBalancer 10.8.7.24 <pending> 80:30539/TCP 5s
The <pending> EXTERNAL-IP will update after a few minutes (run the command
again to check). Once it’s there, copy the IP and paste it into a new browser
tab. This should welcome you with a “Hello from NFS” message. In order to
enable file browsing, we need to delete the index.html file in the pod.
Determine the pod name using the first command listed below and adjust the
second command accordingly.
kubectl get pods -n argo
kubectl exec http-fileserver-XXXXXXXX-YYYYY -n argo -- rm /usr/share/nginx/html/index.html
Warning: anyone can now access these files
This IP is now accessible from anywhere in the world, and therefore also your files (mind: there are charges for outgoing bandwidth). Please delete the service again once you have finished downloading your files.
kubectl delete svc/http-fileserver -n argoRun the
kubectl expose deploymentcommand to expose it again.
Remember to delete your workflow again to avoid additional charges.
argo delete -n argo @latest
Key Points
CMS Open Data workflows can be run in a commercial cloud environment using modern tools
Downloading data using the cernopendata-client
Overview
Teaching: 10 min
Exercises: 15 minQuestions
How can I download data from the Open Data portal?
Should I stream the data from CERN or have them available locally?
Objectives
Understand basic usage of cernopendata-client
The cernopendata-client is a tool that has recently become available, which allows you to access records from the CERN Open Data portal via the command line.
cernopendata-client-go
is a light-weight implementation of this with a particular focus on usage “in
the cloud”. Additionally, it allows you to download records in parallel.
We will be using cernopendata-client-go in the following. Also, mind that
you can also execute these commands on your local computer, but they will also
work in the CloudShell (which is Linux_x86_64).
Getting the tool
You can download the latest release from
GitHub
For use on your own computer, download the corresponding binary archive for
your processor architecture and operating system. If you are on MacOS Catalina
or later, you will have to right-click on the extracted binary, hold CTRL
when clicking on “Open”, and confirm again to open. Afterwards, you should
be able to execute the file from the command line.
The binary will have a different name depending on your operating system and architecture. Execute it to get help and get more help by using the available sub-commands:
./cernopendata-client-go --help
./cernopendata-client-go list --help
./cernopendata-client-go download --help
As you can see from the releases page, you can also use docker instead of downloading the binaries:
docker run --rm -it clelange/cernopendata-client-go --help
Listing files
When browsing the CERN Open Data portal, you will see from the URL that every
record has a number. For example,
Analysis of Higgs boson decays to two tau leptons using data and simulation of events at the CMS detector from 2012
has the URL
http://opendata.web.cern.ch/record/12350. The record ID is therefore 12350.
You can list the associated files as follows:
docker run --rm -it clelange/cernopendata-client-go list -r 12350
This yields:
http://opendata.cern.ch/eos/opendata/cms/software/HiggsTauTauNanoAODOutreachAnalysis/HiggsTauTauNanoAODOutreachAnalysis-1.0.zip
http://opendata.cern.ch/eos/opendata/cms/software/HiggsTauTauNanoAODOutreachAnalysis/histograms.py
http://opendata.cern.ch/eos/opendata/cms/software/HiggsTauTauNanoAODOutreachAnalysis/plot.py
http://opendata.cern.ch/eos/opendata/cms/software/HiggsTauTauNanoAODOutreachAnalysis/skim.cxx
Downloading files
You can download full records in an analoguous way:
docker run --rm -it clelange/cernopendata-client-go download -r 12350
By default, these files will end up in a directory called download, which
contains directories of the record ID (i.e. ./download/12350 here).
Creating download jobs
As mentioned in the introductory slides, it is advantageous to download the
desired data sets/records to cloud storage before processing them.
With the cernopendata-client-go this can be achieved by creating Kubernetes
Jobs.
A job to download a record making use of the volume we have available could look like this:
# job-data-download.yaml
apiVersion: batch/v1
kind: Job
metadata:
name: job-data-download
spec:
backoffLimit: 2
ttlSecondsAfterFinished: 100
template:
spec:
volumes:
- name: volume-opendata
persistentVolumeClaim:
claimName: nfs-<NUMBER>
containers:
- name: pod-data-download
image: clelange/cernopendata-client-go
args: ["download", "-r", "12351", "-o", "/opendata"]
volumeMounts:
- mountPath: "/opendata/"
name: volume-opendata
restartPolicy: Never
Again, please adjust the <NUMBER>.
You can create (submit) the job like this:
kubectl apply -n argo -f job-data-download.yaml
The job will create a pod, and you can monitor the progress both via the pod and the job resources:
kubectl get pod -n argo
kubectl get job -n argo
The files are downloaded to the storage volume, and you can see them through the fileserver as instructed in the previous episode or through the storage pod:
kubectl exec pod/pv-pod -n argo -- ls /mnt/data/
Challenge: Download all records needed
In the following, we will run the Analysis of Higgs boson decays to two tau leptons using data and simulation of events at the CMS detector from 2012 , for which 9 different records are listed on the webpage. We only downloaded the GluGluToHToTauTau dataset so far. Can you download the remaining ones using
Jobs?
Solution: Download all records needed
In principle, the only thing that needs to be changed is the record ID argument. However, resources need to have a unique name, which means that the job name needs to be changed or the finished job(s) have to be deleted.
Key Points
It is usually of advantage to have the data where to CPU cores are.
Running large-scale workflows
Overview
Teaching: 10 min
Exercises: 10 minQuestions
How can I run more than a toy workflow?
Objectives
Run a workflow with several parallel jobs
Autoscaling
Google Kubernetes Engine allows you to configure your cluster so that it is automatically rescaled based on pod needs.
When creating a pod you can specify how much of each reasource a container needs. More information on compute resources can be found on Kubernetes pages. This information is then used to schedule your pod. If there is no node matching your pod’s requirement then it has to wait until some more pods are terminated or new nodes is added.
Cluster autoscaler keeps an eye on the pods scheduled and checks if adding a new node, similar to the other in the cluster, would help. If yes, then it resizes the cluster to accommodate the waiting pods.
Cluster autoscaler also scales down the cluster to save resources. If the autoscaler notices that one or more nodes are not needed for an extended period of time (currently set to 10 minutes) it downscales the cluster.
To configure the autoscaler you simply specify a minimum and maximum for the node pool. The autoscaler then periodically checks the status of the pods and takes action accordingly. You can set the configuration either with the gcloud command-line tool or via the dashboard.
Deleting pods automatically
Argo allows you to describes the strategy to use when deleting completed pods. The pods are deleted automatically without deleting the workflow. Define one of the following strategies in your Argo workflow under the field spec:
spec:
podGC:
# pod gc strategy must be one of the following
# * OnPodCompletion - delete pods immediately when pod is completed (including errors/failures)
# * OnPodSuccess - delete pods immediately when pod is successful
# * OnWorkflowCompletion - delete pods when workflow is completed
# * OnWorkflowSuccess - delete pods when workflow is successful
strategy: OnPodSuccess
Scaling down
Occasionally, the cluster autoscaler cannot scale down completely and extra nodes are left hanging behind. Some situations like those can be found documented here. Therefore it is useful to know how to manually scale down your cluster.
Click on your cluster, listed at Kubernetes Engine - Clusters. Scroll down to the end of the page where you will find the Node pools section. Clicking on your node pool will take you to its details page.
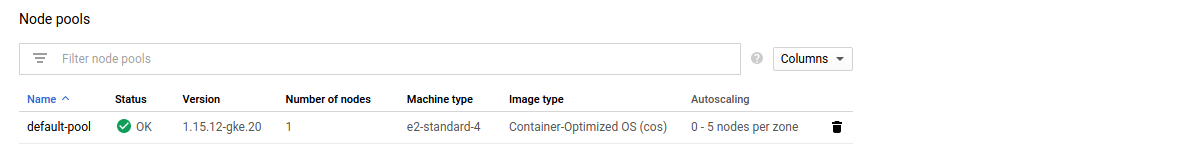
In the upper right corner, next to EDIT and DELETE you’ll find RESIZE.
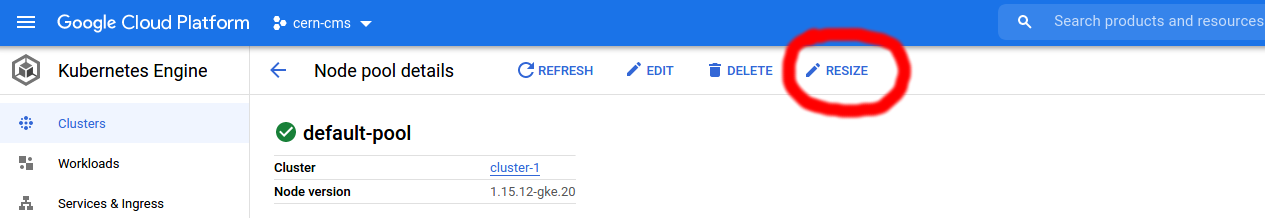
Clicking on RESIZE opens a textfield that allows you to manually adjust the number of pods in your cluster.
Key Points
Argo Workflows on Kubernetes are very powerful.
Building a Docker image on GCP
Overview
Teaching: 10 min
Exercises: 30 minQuestions
How do I push to a private registry on GCP?
Objectives
Build a Docker image on GCP and push it to the private container registry GCR.
Creating your Docker image
The first thing to do when you want to create your Dockerfile is to ask yourself what you want to do with it. Our goal here is to run a physics analysis with the ROOT framework. To do this our Docker container must contain ROOT as well as all the necessary dependencies it requires. Fortunately there are already existing images matching this, such as rootproject/root-conda:6.18.04. We will use this image as our base.
Next we need our source code. We will git clone this into our container from the Github repository https://github.com/awesome-workshop/payload.git.
To save time we will compile the source code. The other option is to add the source code to our image without compiling it. However by compiling the code at this step, we avoid having to do it over and over again as our workflow runs multiple replicas of our container.
In in the CloudShell, create a file compile.sh with the following contents
#!/bin/bash
# Compile executable
echo ">>> Compile skimming executable ..."
COMPILER=$(root-config --cxx)
FLAGS=$(root-config --cflags --libs)
$COMPILER -g -O3 -Wall -Wextra -Wpedantic -o skim skim.cxx $FLAGS
Make it executable with
chmod +x compile.sh
Finally create a Dockerfile that executes everything mentioned above. Create the file Dockerfile with the following contents
# The Dockerfile defines the image's environment
# Import ROOT runtime
FROM rootproject/root-conda:6.18.04
# Argument available during build-time
ARG HOME=/home
# Set up working directory
WORKDIR $HOME
# Clone source code
RUN git clone https://github.com/awesome-workshop/payload.git
WORKDIR $HOME/payload
# Compile source code
ADD compile.sh $HOME/payload
RUN ./compile.sh
Building your Docker image
A Docker image can be built with the docker build command:
docker build -t [IMAGE-NAME]:[TAG] .
If you do not specify a container registry your image will be pushed to DockerHub, the default registry. We would like to push our images to the GCP container registry. To do so we need to name it gcr.io/[PROJECT-ID]/[IMAGE-NAME]:[TAG]
, where [PROJECT-ID] is your GCP project ID. You can find your ID by clicking your project in the top left corner, or in the cloud shell prompt. In our case the ID is cern-cms. The hostname gcr.io tells the client to push it to the GCP registry.
[IMAGE-NAME] and [TAG] are up to you to choose, however a rule of thumb is to be as descriptive as possible. Due to Docker naming rules we aslo have to keep them lowercase. One example of how the command could look like is:
docker build -t gcr.io/cern-cms/root-conda-<NUMBER>:higgstautau .
Replace <NUMBER> with the number for the login credentials you received.
Choose a unique name
Note that all attendants of this workshop are sharing the same registry! If you fail to choose a unique name and tag combination you risk overwriting the image of a fellow attendee. Naming your image with the number of your login credentials is a good way to keep that from happening.
Adding your image to the container registry
Before you push or pull images on GCP you need to configure Docker to use the gcloud command-line tool to autheticate with the registry. Run the following command
gcloud auth configure-docker
This will list some warnings and instructions, which you can ignore for this tutorial. To push the image run
docker push gcr.io/cern-cms/root-conda-<NUMBER>:higgstautau
View your images
You can view images hosted by the container registry via the cloud console, or by visiting the image’s registry name in your web browser at
http://gcr.io/cern-cms/root-conda-<NUMBER>.
Cleaning up your private registry
To avoid incurring charges to your GCP account you can remove your Docker image from the container registry. Do not execute this command right now, we want to use our container in the next step.
When it is time to remove your Docker image, execute the following command:
gcloud container images delete gcr.io/cern-cms/root-conda-<NUMBER>:higgstautau --force-delete-tags
Key Points
GCP allows you to store your Docker images in your own private container registry.
Getting real
Overview
Teaching: 5 min
Exercises: 20 minQuestions
How can I now use this for real?
Objectives
Get an idea of a full workflow with argo
So far we’ve run smaller examples, but now we have everything at hands to run physics analysis jobs in parallel.
Download the workflow with the following command:
curl -OL https://raw.githubusercontent.com/cms-opendata-workshop/workshop-payload-kubernetes/master/higgs-tau-tau-workflow.yaml
The file will look something like this:
apiVersion: argoproj.io/v1alpha1
kind: Workflow
metadata:
generateName: parallelism-nested-
spec:
arguments:
parameters:
- name: files-list
value: |
[
{"file": "GluGluToHToTauTau", "x-section": "19.6", "process": "ggH"},
{"file": "VBF_HToTauTau", "x-section": "1.55", "process": "qqH"},
{"file": "DYJetsToLL", "x-section": "3503.7", "process": "ZTT"},
{"file": "TTbar", "x-section": "225.2", "process": "TT"},
{"file": "W1JetsToLNu", "x-section": "6381.2", "process": "W1J"},
{"file": "W2JetsToLNu", "x-section": "2039.8", "process": "W2J"},
{"file": "W3JetsToLNu", "x-section": "612.5", "process": "W3J"},
{"file": "Run2012B_TauPlusX", "x-section": "1.0", "process": "dataRunB"},
{"file": "Run2012C_TauPlusX", "x-section": "1.0", "process": "dataRunC"}
]
- name: histogram-list
value: |
[
{"file": "GluGluToHToTauTau", "x-section": "19.6", "process": "ggH"},
{"file": "VBF_HToTauTau", "x-section": "1.55", "process": "qqH"},
{"file": "DYJetsToLL", "x-section": "3503.7", "process": "ZTT"},
{"file": "DYJetsToLL", "x-section": "3503.7", "process": "ZLL"},
{"file": "TTbar", "x-section": "225.2", "process": "TT"},
{"file": "W1JetsToLNu", "x-section": "6381.2", "process": "W1J"},
{"file": "W2JetsToLNu", "x-section": "2039.8", "process": "W2J"},
{"file": "W3JetsToLNu", "x-section": "612.5", "process": "W3J"},
{"file": "Run2012B_TauPlusX", "x-section": "1.0", "process": "dataRunB"},
{"file": "Run2012C_TauPlusX", "x-section": "1.0", "process": "dataRunC"}
]
entrypoint: parallel-worker
volumes:
- name: task-pv-storage
persistentVolumeClaim:
claimName: nfs-<NUMBER>
templates:
- name: parallel-worker
inputs:
parameters:
- name: files-list
- name: histogram-list
dag:
tasks:
- name: skim-step
template: skim-template
arguments:
parameters:
- name: file
value: ""
- name: x-section
value: ""
withParam: ""
- name: histogram-step
dependencies: [skim-step]
template: histogram-template
arguments:
parameters:
- name: file
value: ""
- name: process
value: ""
withParam: ""
- name: merge-step
dependencies: [histogram-step]
template: merge-template
- name: plot-step
dependencies: [merge-step]
template: plot-template
- name: fit-step
dependencies: [merge-step]
template: fit-template
- name: skim-template
inputs:
parameters:
- name: file
- name: x-section
script:
image: gcr.io/cern-cms/root-conda-002:higgstautau
command: [sh]
source: |
LUMI=11467.0 # Integrated luminosity of the unscaled dataset
SCALE=0.1 # Same fraction as used to down-size the analysis
mkdir -p $HOME/skimm
./skim root://eospublic.cern.ch//eos/opendata/cms/derived-data/AOD2NanoAODOutreachTool/.root $HOME/skimm/-skimmed.root $LUMI $SCALE
ls -l $HOME/skimm
cp $HOME/skimm/* /mnt/vol
volumeMounts:
- name: task-pv-storage
mountPath: /mnt/vol
resources:
limits:
memory: 2Gi
requests:
memory: 1.7Gi
cpu: 750m
- name: histogram-template
inputs:
parameters:
- name: file
- name: process
script:
image: gcr.io/cern-cms/root-conda-002:higgstautau
command: [sh]
source: |
mkdir -p $HOME/histogram
python histograms.py /mnt/vol/-skimmed.root $HOME/histogram/-histogram-.root
ls -l $HOME/histogram
cp $HOME/histogram/* /mnt/vol
volumeMounts:
- name: task-pv-storage
mountPath: /mnt/vol
resources:
limits:
memory: 2Gi
requests:
memory: 1.7Gi
cpu: 750m
- name: merge-template
script:
image: gcr.io/cern-cms/root-conda-002:higgstautau
command: [sh]
source: |
hadd -f /mnt/vol/histogram.root /mnt/vol/*-histogram-*.root
ls -l /mnt/vol
volumeMounts:
- name: task-pv-storage
mountPath: /mnt/vol
resources:
limits:
memory: 2Gi
requests:
memory: 1.7Gi
cpu: 750m
- name: plot-template
script:
image: gcr.io/cern-cms/root-conda-002:higgstautau
command: [sh]
source: |
SCALE=0.1
python plot.py /mnt/vol/histogram.root /mnt/vol $SCALE
ls -l /mnt/vol
volumeMounts:
- name: task-pv-storage
mountPath: /mnt/vol
- name: fit-template
script:
image: gcr.io/cern-cms/root-conda-002:higgstautau
command: [sh]
source: |
python fit.py /mnt/vol/histogram.root /mnt/vol
ls -l /mnt/vol
volumeMounts:
- name: task-pv-storage
mountPath: /mnt/vol
Adjust the workflow as follows:
- Replace
gcr.io/cern-cms/root-conda-002:higgstautauin all places with the name of the image you created - Adjust
claimName: nfs-<NUMBER>
Then execute the workflow and keep your thumbs pressed:
argo submit -n argo --watch higgs-tau-tau-workflow.yaml
Good luck!
Key Points
Argo is a powerful tool for running parallel workflows
Cleaning up
Overview
Teaching: 3 min
Exercises: 2 minQuestions
How can I delete my cluster and disks?
Objectives
Know what needs to be deleted to not be charged.
Two things need to be done:
- In the Cluster Overview page, click on the trashbin icon next to your cluster and confirm.
- In the Disks Overview, delete the disk you created
Key Points
The cluster and disks should be deleted if not needed anymore.